Coronavirus SARS-CoV-2 and the COVID-19 Pandemic: What is it and how does it spread?
- Jump To:
- References
Coronaviruses are a large group of viruses known to cause mild to severe acute respiratory illnesses (SARs) in humans. The particular virus that is the cause of the 2019-2020 novel Coronavirus outbreak is the SARS-CoV-2 virus, which results in the COVID-19 illness and is the cause of the COVID-19 global pandemic. [SARS-CoV-2 is technically the virus, and COVID-19 is the illness; the terms are often used interchangeably].
Coronavirus is most often transmitted through viral shedding, where the virus is transmitted from one individual to another (community transfer) or through environmental contamination. Viruses are not “alive” as bacteria are. They are referred to as virions or “viral particles” and are essentially nucleic acid (RNA or DNA) surrounded by a protein-lipid-membrane coat, which infects a mammalian host cell and commandeers the biochemical machinery of that cell to produce the components to make new virions. There has been much speculation about possible animal origins of SARS-CoV-2 given that the RNA shows some similarity to SARS-CoV-1 (80% similarity) and an even higher similarity to SARS-CoV virus isolated from pangolin (91%) and bats (96%) [1].
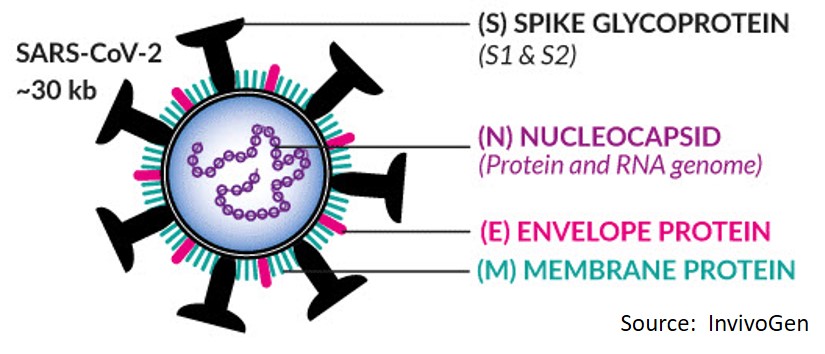
Coronavirus-2 (SARS-CoV-2, COVID-19) is an RNA virus surrounded by a coat that has prominent “spike proteins” protruding from its surface. These spike proteins (glycoproteins) are the main molecule that initiates attachment to receptors on human cells. Its genome consists of single stranded RNA that is complexed with nucleocapsid protein (N protein), which helps with RNA folding. Envelope (E) protein and membrane protein (M) form the bulk of the remaining capsid shell proteins and assist with the fusion of host and viral membranes required for host infection.
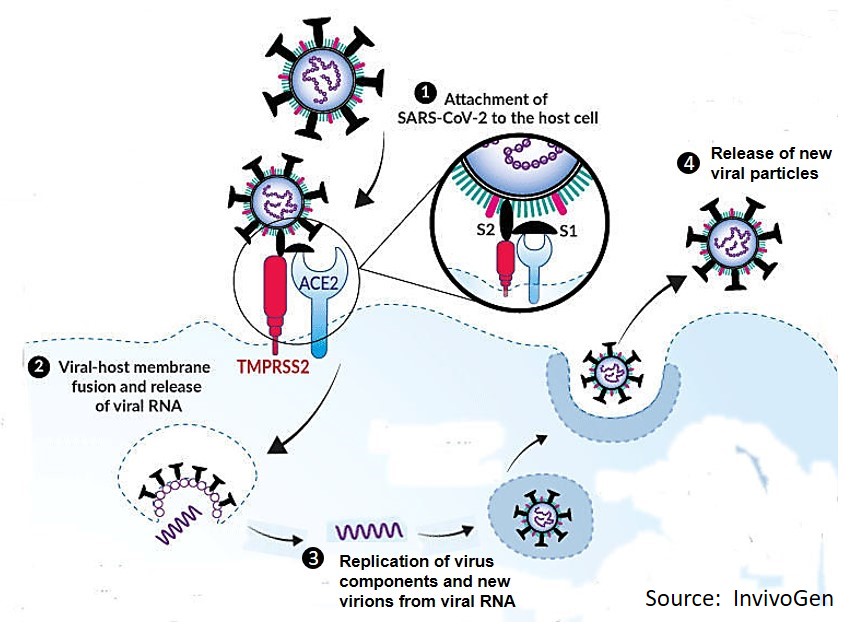
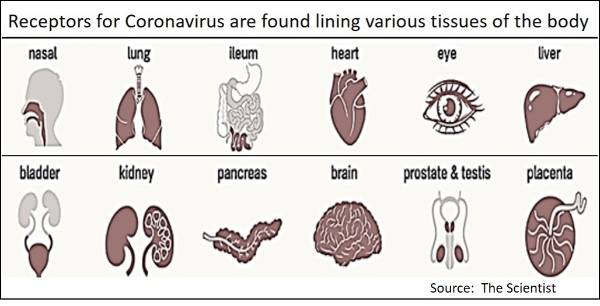
The receptor on human host cells for the SARS-CoV-2 virus is a transmembrane protein, ACE2 (i.e., angiotensin converting enzyme 2). The human ACE2 receptor proteins are on the surface of many types of human cells including the epithelial lining of the lung, intestine, liver, kidney, heart and other tissues. The ACE2 receptor serves as the main binding receptor for the viral spike protein and helps mediate entry into human cells that end up serving as host cells for viral replication. Receptors often extend through a cell membrane (i.e., transmembrane), whereby binding externally at the receptor initiates biochemical activity at the other end of the molecule inside the cell. Several other host transmembrane enzyme proteins (proteases) also participate during infection (TMPRSS2, CTSL) in which there is a dual binding activation, first ACE2 binding to Spike protein and then binding and proteolytic activity of Spike by TMPRSS2 (or CTSL). Binding of the viral spike protein to the host membrane ACE2 receptor is the initial binding event and is further assisted by a proteolytic membrane protein (TMPRSS2). The result of this “tag team” receptor action at the cell membrane results in the virus coat membrane fusing with the human host cell membrane. This releases the virus genome (RNA) into the host cell cytoplasm, taking over the host cells’ biochemical machinery to direct assembly and packaging of new viral particles that are released by “budding.”
Although receptors for the SARS-CoV-2 virus are found in the lung, intestine, liver, kidney, heart and other tissues, it is commonly recognized the primary site of human infection is in the lung. One study has shown children do not yet have high level expression of ACE2 in their lungs, and this may explain why they are not infected as readily as adults. Infection of the intestinal tract epithelium because of ACE2 receptors found there may explain why a high proportion of patients experience diarrhea as one of their symptoms, although there have been disputed claims of whether active virus, or simply the SARS-CoV-19 RNA, has been isolated from feces. The isolation of active virus from feces would be significant because of the potential for hand-to-mouth spread if sanitary practices are not strictly adhered to after such diarrheal episodes. A recent article discussed the potential for “toilet spray” aerosolization from flushing of diarrheal episodes resulting in 40-60% of aerosolized viruses rising above the toilet seat [2]. Further, widespread dissemination could occur if sewage treatment centers are overwhelmed with excess rainstorm runoff water and subsequently release untreated sewage that could contain active virus particles. This is the reason health departments of coastal towns examine water for contamination by “fecal coliforms” to determine the quality of water for bathers and the suitability of water quality for shellfish harvesting from shoreline clam beds. The COVID-19 pandemic has broadened the potential for examining sewage treatment water, not only for bacterial pathogens but also for use as an epidemiological barometer of the extent of viral presence in a community served by the treatment facility [3].
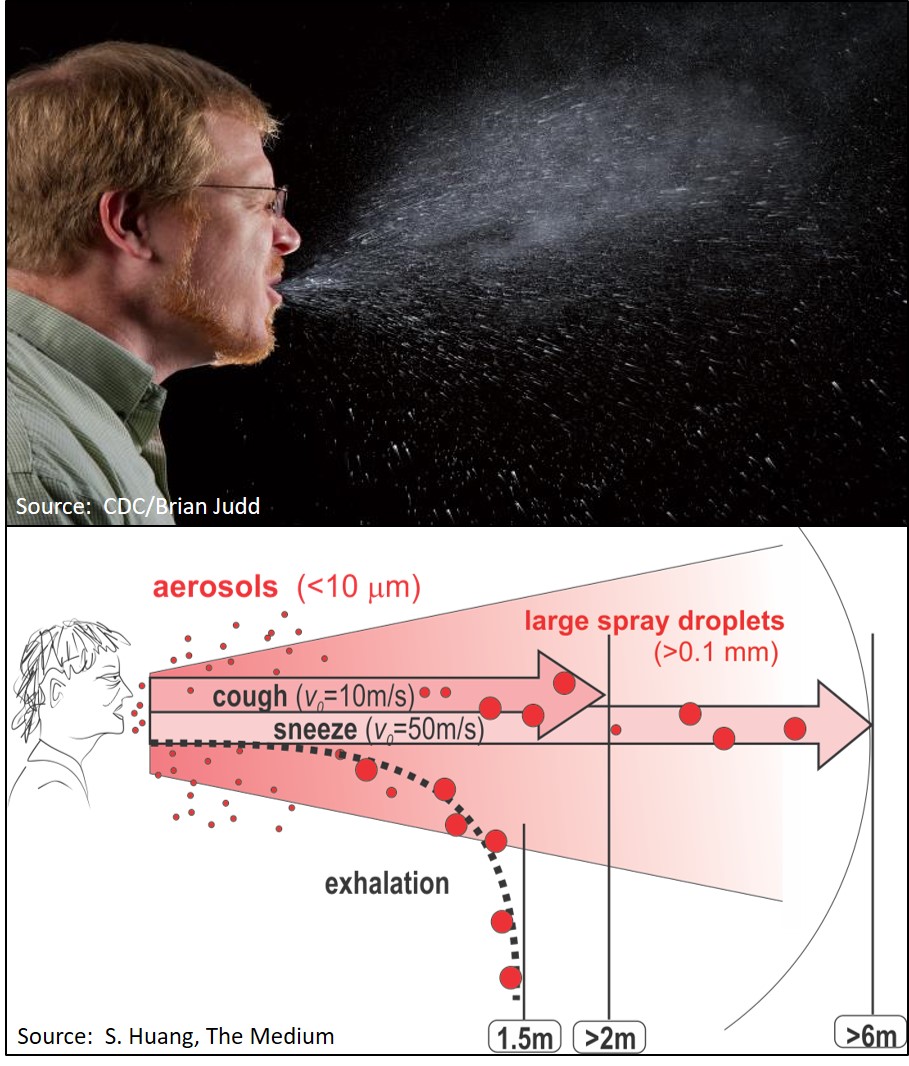
Experts at CDC believe the majority of SARS-CoV-2 infections are spread and acquired through respiratory droplets, whereby the virus is aerosolized upon sneezing, coughing and even by simply talking or breathing. Violent sneezes have been shown to generate aerosolized droplets that can travel as far as 23-27 feet and further, depending on the size of the particle [4,5]. Small aerosolized droplets can stay suspended without evaporating for longer periods in humid climates. The distance aerosolized particles can travel from a cough or sneeze is significant given the number of viral particles that could be present in sputum or nasal discharge. Quantitative analysis of viral loads found levels of approximately 2-8 log10 (102-108) viral particles per ml (depending on the virus examined) of sputum, which demonstrated higher viral loads than swabs dunked in sputum (“dunked sputum”) or from nose and throat swabs (NTS). Other studies quantifying RNA have shown high levels in sputum (106-1011 RNA/ml) vs nasal (106-109 RNA/swab) or throat (106-109 RNA/swab) swabs [6]. It is easy to speculate if billions of viral particles are generated by a single infected person/animal, low incidence mutational changes in the RNA genome that reflect the amino acids of the receptor binding domain, can result in a sequence that is even better suited to binding a specific type of host cell. This would make for a greater likelihood of infectivity or can even change the specificity to a different species.
The manner in which new virus particles are released is not like the “burst size” of lytic bacteriophage, which are released from infected bacteria by lysing or “bursting” out from the host cell and killing them. Newly formed COVID-19 virus particles are released from infected cells by continuously “budding” new virions from the surface of infected host cells without killing them. Based on tissue culture cell analysis, it takes about 10 minutes to infect a host cell, around 10 hours to generate a new virion particle, and each host cell can release as many as 1,000 virus particles over the duration of the infection [7]. This explains why sputum and nasal discharge have such high levels of virus particles and even the gentle exhale during talking can create aerosolized droplets containing COVID-19 virus. A more violent discharge (sneeze, cough) can create an array of droplets of different sizes, and smaller ones can travel further via air currents before landing on surfaces or being inhaled by other humans. Investigations in two hospitals in Wuhan, China, found infectious COVID-19 RNA in the air in as small as sub-micron droplet size and initiated a program to disinfect the air where COVID-19 patients had been [8]. The COVID-19 virus can remain infectious for numerous hours after settling on various surfaces and the ability to infect has been examined in reference to “half-life” of infectivity. Large scale, area-wide fogging systems to permeate entire rooms have been used to disinfect hospital rooms that once held Coronavirus patients, and high-intensity ultraviolet light systems have been installed in ductwork to sanitize aerosolized virus particles carried in the air distributed by ventilation systems.
Once infected, it takes about 3 days for a person to become infectious (i.e., “latent period”) and then that person will be infectious for about 4 days before showing symptoms. It is during this asymptomatic infectious period where an infected person can be infecting other people if there are no distancing or if mask wearing practices are not adopted. Infectivity and spread of infection often are referred to by the “effective reproduction number” (R0, “R naught” or “R zero”). An R0 = 1 indicates for every 1 person infected, it is transmitted to 1 other person. An R0<1 suggests spreading can be controlled; an R0>1 indicates spreading of an infection among the population. With an R0=4, it is expected the number of cases will approximately double every ~3 days and quadruple every ~7 days. The latency and infectious period can be variable; an epidemiological analysis of the outbreak in Wuhan, China, suggests the latency incubation period was 5.1 days and 97.5% of those that developed symptoms did so within 11.5 days and 1% of the cases did not show symptoms until 14 days. So, the longer the infectious period is maintained without symptoms, the greater the number of people who can become infected, especially when there are no countermeasures (masks, distancing) being practiced. This is where testing and availability of test results can help by identifying who is infected (and not yet showing symptoms), so they may be quarantined instead of contributing more to the spread of the virus.
There has been much discussion about infectivity by virus particles left behind on various types of surfaces (door knobs, tables, desks, packages in supermarket, push buttons, cell phones, etc.) and studies have shown COVID-19 virus may last for some limited time on such surfaces. Since viruses are not “alive,” the timeframe of their infectivity has more to do with denaturation of the outer protein/coat components, so they cannot interact with their human host cell receptors and possible irreversible binding of the virus proteins to surfaces. The “decay” of infective titers of SARS-CoV-1 vs. SARS-CoV-2 have been compared on various surfaces and have shown titers diminish greatest on copper and slowest on plastic surfaces (copper > cardboard > stainless steel > plastic). Also, the infectivity of virus particles can persist for hours when aerosolized [9].

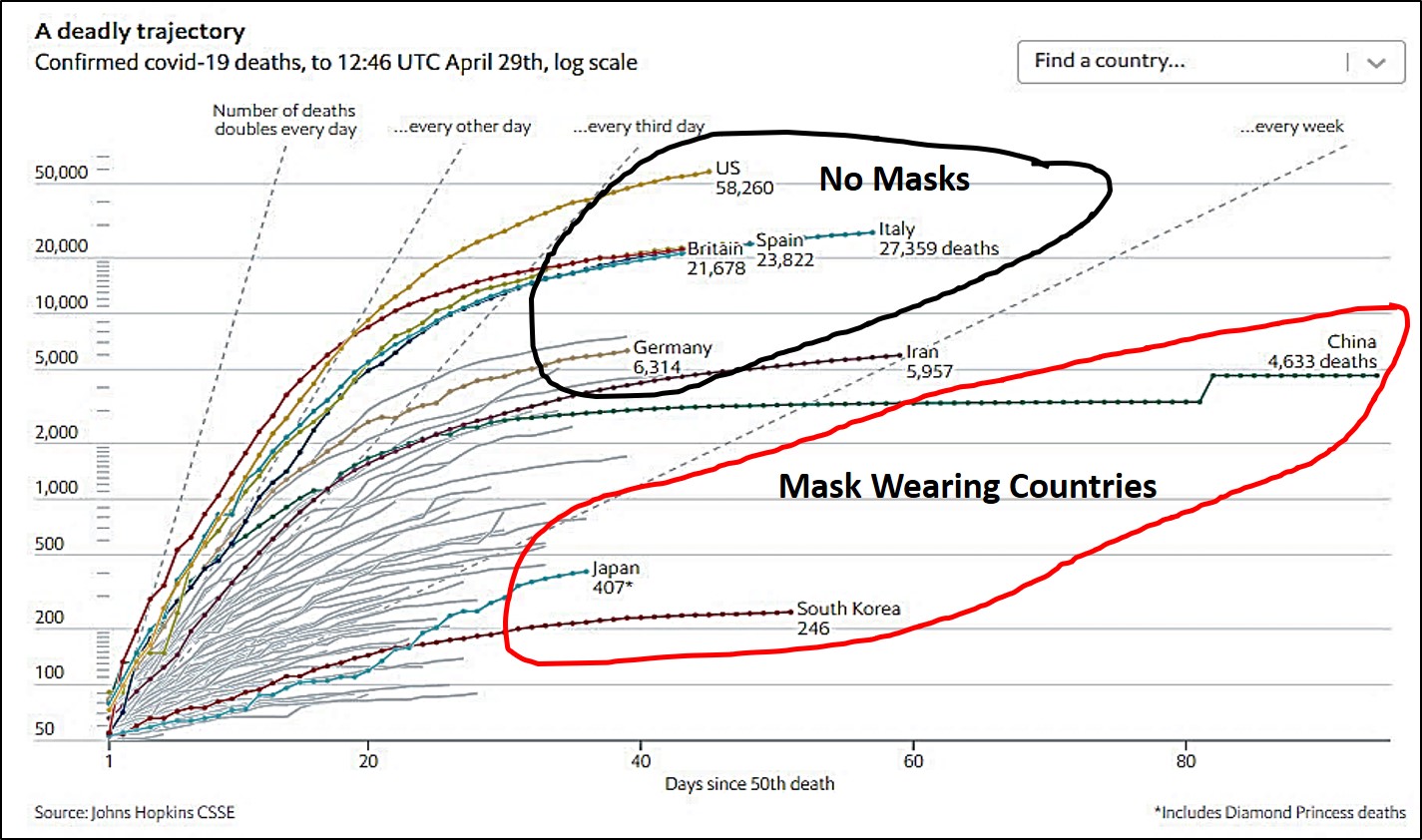
The potential infectiveness of airborne/aerosolized COVID-19 particles underlies the importance of wearing masks to prevent widespread expulsion of virus particles by infected asymptomatic, presymptomatic or weakly symptomatic people, or inhalation uptake by yet uninfected people. In certain parts of the world, people are more accustomed to using face masks, if not for protection from infectious disease, then often because of widespread air pollution. People in Western nations are not as accustomed to wearing masks as those in some Asian nations, and this may have resulted in holding the spread of virus in check or at least providing a lower trajectory of increase in the number of cases among those cultures. The importance of wearing face masks is borne out in the most recent CDC recommendations.
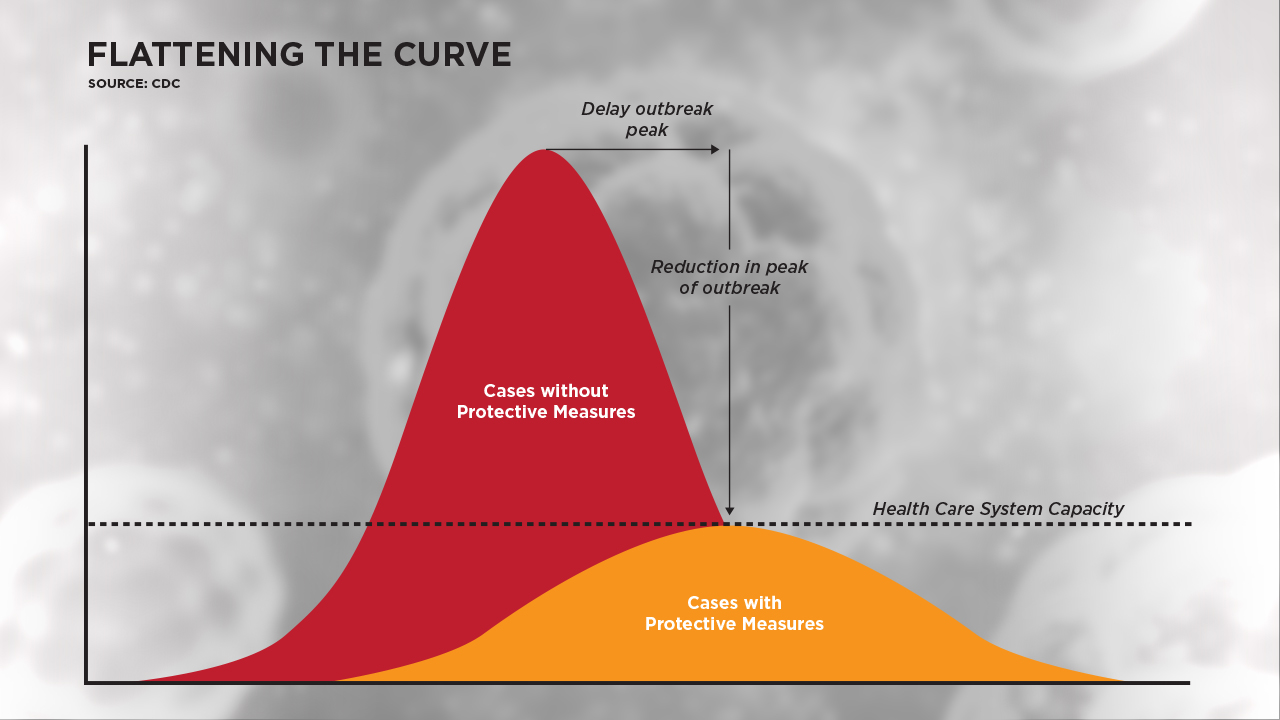
The intention of wearing masks and other preventive measures (i.e., social distancing) is to reduce the spread of virus (i.e., R0 ≤ 1.0) so as not to overburden the healthcare system in regard to unrelenting burden on healthcare workers, their supply of personal protective equipment and availability of intensive care unit beds. Reducing the trajectory of an outbreak is often referred to as “flattening the curve.” Putting such measures into practice may not completely control the outbreak, but they can lessen the intensity of the virus burden on the healthcare system so it can better work with the cases of illness.
References
- Bar-On, Y.M.; Flamholz, A.; Phillips, R.; Milo, R. SARS-CoV-2 (COVID-19) by the numbers. eLife 2020, 9, e57309.
- Li, Y.-Y.; Wang, J.-X.; Chen, X. Can a toilet promote virus transmission? From a fluid dynamics perspective. Phys. Fluids 2020, 32, 065107.
- Randazzo, W.; Cuevas-Ferrando, E.; Sanjuan, R.; Domingo-Calap, P.; Sanchez, G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. medRxiv 2020, 2020.2004.2023.20076679.
- Bourouiba, L. Turbulent gas clouds and respiratory pathogen emissions: potential implications for reducing transmission of COVID-19. Jama 2020.
- Huang, S. COVID-19: Why we should all wear masks - There is new scientific rationale. In Medium, 2020; Vol. 2020.
- Branche, A.R.; Walsh, E.E.; Formica, M.A.; Falsey, A.R. Detection of respiratory viruses in sputum from adults by use of automated multiplex PCR. J. Clin. Microbiol. 2014, 52, 3590-3596.
- Lauer, S.A.; Grantz, K.H.; Bi, Q.; Jones, F.K.; Zheng, Q.; Meredith, H.R.; Azman, A.S.; Reich, N.G.; Lessler, J. The incubation period of Coronavirus Disease 2019 (COVID-19) from publicly reported confirmed cases: Estimation and application. Ann. Intern. Med. 2020, 172, 577-582.
- Liu, Y.; Ning, Z.; Chen, Y.; Guo, M.; Liu, Y.; Gali, N.K.; Sun, L.; Duan, Y.; Cai, J.; Westerdahl, D.; Liu, X.; Xu, K.; Ho, K.-F.; Kan, H.; Fu, Q.; Lan, K. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature 2020.
- Van Doremalen, N.; Bushmaker, T.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; Lloyd-Smith, J.O.; De Wit, E.; Munster, V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. New Eng. J. Med. 2020, 382, 1564-1567.